Canine Array Comparative Genomic Hybridization (aCGH)
Many human and canine cancers exhibit highly comparable clinical presentation and progression, and as with many human cancers, canine tumors also demonstrate recurrent chromosome aberrations. In both species, it has been observed that even in tumors presenting with apparently similar histopathology, the pattern of chromosome aberrations may be distinctly different. This indicates that the tumors represent different genetic subgroups of the same cancer, and may also demonstrate disparity in their subsequent clinical behavior. A detailed knowledge of the pattern of chromosome aberrations in genetically heterogeneous tumors may therefore improve existing methods for diagnosis, prognosis and the selection of appropriate therapy.
Patterns of recurrent chromosome aberrations may be characterized using a range of techniques. Amongst these, comparative genomic hybridization (CGH) analysis allows a genome-wide survey of chromosome imbalances in a single experiment. These experiments are now typically performed using microarray analysis, since array-based CGH enables DNA copy number abnormalities to be detected more efficiently and at higher resolution than conventional chromosome-based techniques.
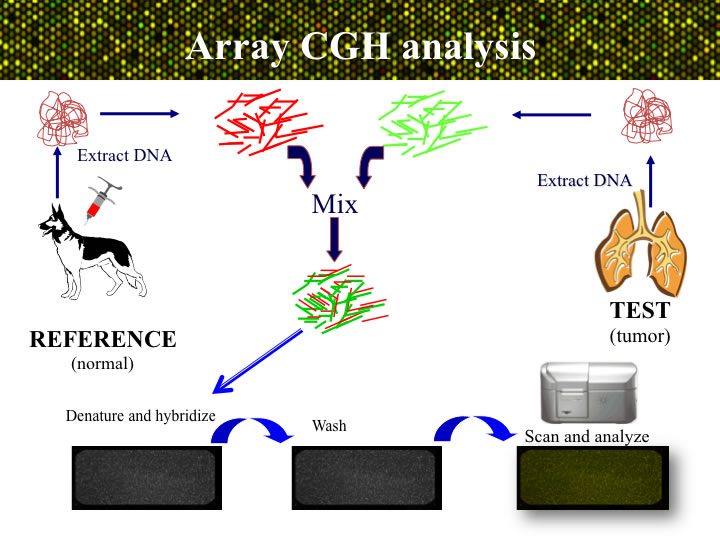
APPROACH: The process of aCGH is summarized in the figure below. Genomic DNA is isolated from the tumor sample sumbitted to the lab, and also from a blood or tissue sample from one or more dogs that do not have cancer. The tumor (or ‘test’) DNA and the healthy (‘reference)’ DNA samples areeach labeled witha different colored fluorophore. The two labeled probes are then combined and hybridized onto the microarray for two days. After a series of stringency washes, the array is scanned to visualize the relative amounts of red (tumor) and green (reference) DNA binding at each genomic target on the microarray. The ratio of these signal intensities indicates the relative amount of DNA representing that target in each of the two DNA samples. Since the reference sample is known to be normal, any deviation from a 1:1 ratio must arise due to a DNA copy number aberration (CNA) of that region in the tumor. The ratio data for each arrayed locus are plotted graphically to generate a genome-wide CGH profile.
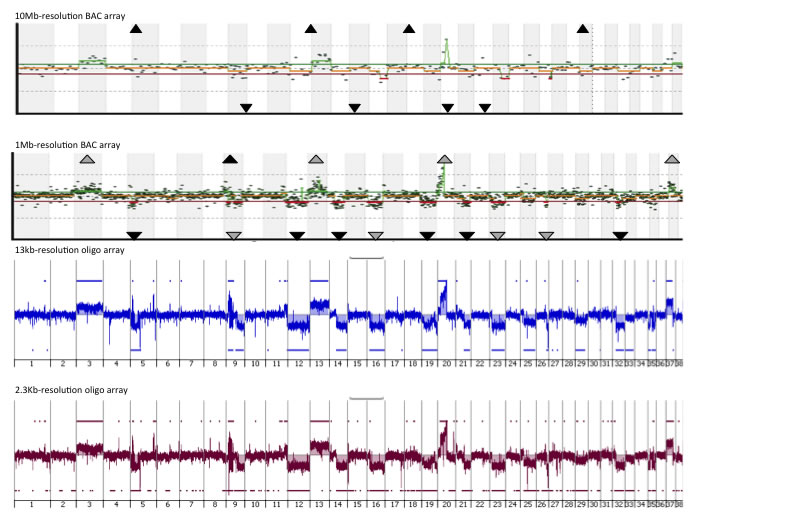
An example of the CGH profiles of the same tumor from arrays with four different resolutions (10Mb and 1Mb arrays, comprising 282 and 2,200 BACs as the probes; 13Kb and 2.3Kb, comprising 180,000 and 1,000,000 oligonucelotides as the probes) are shown below. It is very clear that surveying the genome at higher resolution is able to identify changes that are undetected by lower resolution options.
While arrays generated from BAC clones have the DNA from the BACs printed onto a glass slide, for oligonucleotide arrays the oligonucleotides are designed to represent specific locations in the genome of interest and are then synthesized directly on the surface of a glass slide. The number of probes per unit length of the genome determines the resolution of the array. The physical size of a glass slides allows for multiple arrays (or fields) to be generated if each field is smaller than a slide. For example, Agilent Technologies are ablecurrently to provide four array designs, a single field with 1,000,000 probes, two fields each with 400,000 probes, four fields each with 180,000 probes, or eight fields each with 60,000 probes.
Using aCGH proffiling we detemined the genome-wide DNA copy number status of almost 1,000 canine tumors, representing cases of canine lymphoma, leukemia, central nervous system tumors, osteosarcoma, soft-tissue sarcomas, melanoma, mast cell tumors, urotheloial carcinoma, hemangiosarcoma, histiocytic malignancies.
From these data we have identified recurrent DNA copy changes that are being used to understand the key drivers of these cancers.
We have humanized all the canine data to allow us to compare directly with CGH data derived from the corresponding human cancer. We are using these comparative data to accelerate gene discovery as we seek to determine the drivers and actionable copy nuumbers changes for both canine and human patients.
Typically, aCGH data are further investigated using single locus probe (SLP) FISH analysis on chromosome or interphase nucleus preparations from the tumor samples. This enables the copy number status of individual genomic loci to be evaluated within single cells from the tumor patient, and may also detect balanced structural chromosome rearrangements that are not apparent using aCGH. The SLPs routinely used are derived from bacterial artificial chromossome clones. We have develeoped a panel of over 2,500 canine BAC clones, each of which has been cytogenetically verified, intergrated onto the canine genome, and organized into chromosome-specific sets.
In addition validation of oaCGH data may also be performed using droplet digital PCR.